Azimuthal Averaging#
This guide showcases how to apply Azimuthal Averaging on 2D and 3D fields using UXarray.
Azimuthal mean basics#
An azimuthal average (or azimuthal mean) is a statistical measure that represents the average of a face-centered variable along rings/bands of constant distance from a specified central point. Azimuthal averaging is useful for describing circular/cylindrical features, where fields that strongly depend on distance from a center.
In UXarray, azimuthal averaging is non-conservative. This means that faces are assigned to radial bins (i.e., distance intervals from the central point) based only on the face center coordinate.
# The imports below are used to visualize range rings in this notebook and
# are not needed for routine use of `azimuthal_mean()`.
import math
import operator
from functools import reduce
import cartopy.geodesic as gdyn
import holoviews as hv
import matplotlib.pyplot as plt
import numpy as np
import uxarray as ux
uxds = ux.open_dataset(
"../../test/meshfiles/ugrid/outCSne30/outCSne30.ug",
"../../test/meshfiles/ugrid/outCSne30/outCSne30_vortex.nc",
)
1. Azimuthally averaging an idealized 2D field#
Helper function to draw range rings and visualize azimuthal averaging:#
def hv_range_ring(lon, lat, rad_gcd, n_samples=2000, color="red", line_width=1):
geo = gdyn.Geodesic()
circ_pts = geo.circle(
lon=lon, lat=lat, radius=rad_gcd * 111320, n_samples=n_samples
)
return hv.Path(circ_pts).opts(color=color, line_width=line_width)
Step 1.1: Visualize the global field#
The global field is shaded below using the UxDataArray.plot() accessor. To help visualize azimuthal averaging, the chosen central point is marked with an ‘x’ and rings are drawn at every 10 great-circle degrees from the central point (37°N, 1°E).
# Display the global field
glob_plt = uxds["psi"].plot(
cmap="inferno", periodic_elements="split", title="Global Field", dynamic=True
)
# Mark a center coordinate and draw range rings
lon, lat = 1, 37
glob_plt = glob_plt * hv.Points([(lon, lat)]).opts(
color="lime", marker="x", size=10, line_width=2
)
glob_plt = glob_plt * reduce(
operator.mul,
[hv_range_ring(lon, lat, rr, color="lime") for rr in np.arange(10, 41, 10)],
)
glob_plt
Step 1.2: Compute the azimuthal mean#
Calling .azimuthal_mean() with the arguments below samples every 2° out to 40 great-circle degrees around the central point.
azim_mean_psi, hits = uxds["psi"].azimuthal_mean(
(lon, lat), 40.0, 2, return_hit_counts=True
)
azim_mean_psi
<xarray.DataArray 'psi_azimuthal_mean' (radius: 21)> Size: 168B
array([ nan, 0.98400088, 0.98263337, 0.98076282, 1.03482251,
1.03210027, 0.96639787, 0.94121837, 0.99013955, 1.08111055,
1.08413299, 0.99805499, 0.94448357, 0.90755765, 0.92506048,
0.94187134, 0.93900079, 0.9867494 , 1.00598615, 0.98697273,
0.99283143])
Coordinates:
* radius (radius) float64 168B 0.0 2.0 4.0 6.0 8.0 ... 34.0 36.0 38.0 40.0
Attributes:
azimuthal_mean: True
center_lon: 1.0
center_lat: 37.0
radius_units: degreesStep 1.3: Plot the azimuthal mean#
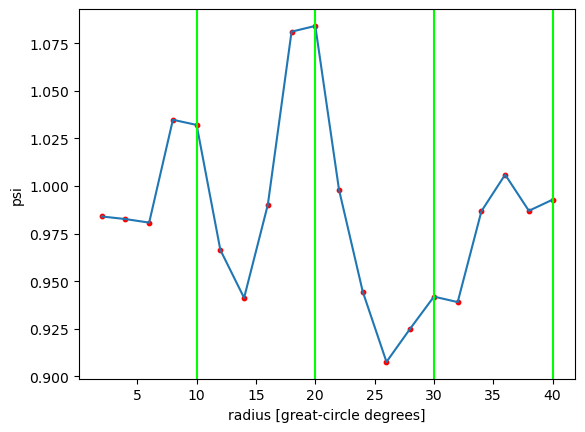
In the plot below, red dots mark where samples were taken at 2° intervals. The green vertical lines correspond to the green rings in the global field plot.
plt.plot(azim_mean_psi["radius"], azim_mean_psi)
plt.scatter(azim_mean_psi["radius"], azim_mean_psi, s=10, color="red")
plt.xlabel("radius [great-circle degrees]")
plt.ylabel("psi")
[plt.axvline(rr, color="lime") for rr in np.arange(10, 41, 10)]
[<matplotlib.lines.Line2D at 0x7ecad7b87b60>,
<matplotlib.lines.Line2D at 0x7ecad7b87e00>,
<matplotlib.lines.Line2D at 0x7ecad7140050>,
<matplotlib.lines.Line2D at 0x7ecad71401a0>]
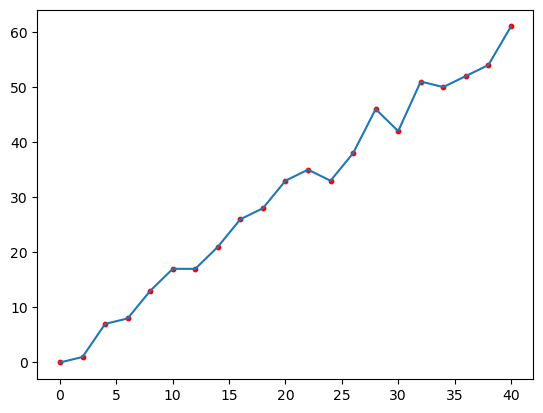
Step 1.4: Inspect the hit count#
The plot below shows the number of face centers that fall within each distance bin. As one would expect on a near-uniformly spaced mesh, the hit count increases linearly with distance.
plt.plot(azim_mean_psi["radius"], hits)
plt.scatter(azim_mean_psi["radius"], hits, s=10, color="red")
<matplotlib.collections.PathCollection at 0x7ecad6ffa5d0>
2: Azimuthally averaging tropical cyclone fields#
A high-resolution (~0.25°) aquaplanet general circulation model permits the development of a handful of strong tropical cyclones (TCs). Because TCs are fairly axisymmetric, azimuthal averaging is useful for transforming their fields into cylindrical coordinates and visualizing features such as their low central pressure and warm core.
clon, clat = 114.54, -17.66
tcds = ux.open_dataset(
"/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc",
"/glade/work/jpan/uxazim_demo/cam.h1i.plev.0013-01-13-00000.nc",
).squeeze()
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:219, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
218 try:
--> 219 file = self._cache[self._key]
220 except KeyError:
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/lru_cache.py:56, in LRUCache.__getitem__(self, key)
55 with self._lock:
---> 56 value = self._cache[key]
57 self._cache.move_to_end(key)
KeyError: [<class 'netCDF4._netCDF4.Dataset'>, ('/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc',), 'r', (('clobber', True), ('diskless', False), ('format', 'NETCDF4'), ('persist', False)), '44f0c3b3-3454-4d02-afe4-db721cac81c6']
During handling of the above exception, another exception occurred:
FileNotFoundError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/uxarray/core/utils.py:33, in _open_dataset_with_fallback(filename_or_obj, chunks, **kwargs)
31 try:
32 # Try opening with xarray's default read engine
---> 33 return xr.open_dataset(filename_or_obj, chunks=chunks, **kwargs)
34 except Exception:
35 # If it fails, use the "netcdf4" engine as backup
36 # Extract engine from kwargs to prevent duplicate parameter error
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/api.py:607, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, create_default_indexes, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
606 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 607 backend_ds = backend.open_dataset(
608 filename_or_obj,
609 drop_variables=drop_variables,
610 **decoders,
611 **kwargs,
612 )
613 ds = _dataset_from_backend_dataset(
614 backend_ds,
615 filename_or_obj,
(...) 626 **kwargs,
627 )
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:771, in NetCDF4BackendEntrypoint.open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, mode, format, clobber, diskless, persist, auto_complex, lock, autoclose)
770 filename_or_obj = _normalize_path(filename_or_obj)
--> 771 store = NetCDF4DataStore.open(
772 filename_or_obj,
773 mode=mode,
774 format=format,
775 group=group,
776 clobber=clobber,
777 diskless=diskless,
778 persist=persist,
779 auto_complex=auto_complex,
780 lock=lock,
781 autoclose=autoclose,
782 )
784 store_entrypoint = StoreBackendEntrypoint()
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:529, in NetCDF4DataStore.open(cls, filename, mode, format, group, clobber, diskless, persist, auto_complex, lock, lock_maker, autoclose)
526 manager = CachingFileManager(
527 netCDF4.Dataset, filename, mode=mode, kwargs=kwargs, lock=lock
528 )
--> 529 return cls(manager, group=group, mode=mode, lock=lock, autoclose=autoclose)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:429, in NetCDF4DataStore.__init__(self, manager, group, mode, lock, autoclose)
428 self._mode = mode
--> 429 self.format = self.ds.data_model
430 self._filename = self.ds.filepath()
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:538, in NetCDF4DataStore.ds(self)
536 @property
537 def ds(self):
--> 538 return self._acquire()
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:532, in NetCDF4DataStore._acquire(self, needs_lock)
531 def _acquire(self, needs_lock=True):
--> 532 with self._manager.acquire_context(needs_lock) as root:
533 ds = _nc4_require_group(root, self._group, self._mode)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/contextlib.py:141, in _GeneratorContextManager.__enter__(self)
140 try:
--> 141 return next(self.gen)
142 except StopIteration:
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:207, in CachingFileManager.acquire_context(self, needs_lock)
206 """Context manager for acquiring a file."""
--> 207 file, cached = self._acquire_with_cache_info(needs_lock)
208 try:
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:225, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
224 kwargs["mode"] = self._mode
--> 225 file = self._opener(*self._args, **kwargs)
226 if self._mode == "w":
227 # ensure file doesn't get overridden when opened again
File src/netCDF4/_netCDF4.pyx:2521, in netCDF4._netCDF4.Dataset.__init__()
File src/netCDF4/_netCDF4.pyx:2158, in netCDF4._netCDF4._ensure_nc_success()
FileNotFoundError: [Errno 2] No such file or directory: '/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc'
During handling of the above exception, another exception occurred:
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:219, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
218 try:
--> 219 file = self._cache[self._key]
220 except KeyError:
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/lru_cache.py:56, in LRUCache.__getitem__(self, key)
55 with self._lock:
---> 56 value = self._cache[key]
57 self._cache.move_to_end(key)
KeyError: [<class 'netCDF4._netCDF4.Dataset'>, ('/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc',), 'r', (('clobber', True), ('diskless', False), ('format', 'NETCDF4'), ('persist', False)), '1673873e-09a2-4300-b0aa-b7c0a07c5f73']
During handling of the above exception, another exception occurred:
FileNotFoundError Traceback (most recent call last)
Cell In[8], line 2
1 clon, clat = 114.54, -17.66
----> 2 tcds = ux.open_dataset(
3 "/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc",
4 "/glade/work/jpan/uxazim_demo/cam.h1i.plev.0013-01-13-00000.nc",
5 ).squeeze()
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/uxarray/core/api.py:414, in open_dataset(grid_filename_or_obj, filename_or_obj, chunks, chunk_grid, use_dual, grid_kwargs, **kwargs)
411 grid_kwargs = {}
413 # Construct a Grid, validate parameters, and correct chunks
--> 414 uxgrid, corrected_chunks = _get_grid(
415 grid_filename_or_obj, chunks, chunk_grid, use_dual, grid_kwargs, **kwargs
416 )
418 # Load the data as a Xarray Dataset
419 ds = _open_dataset_with_fallback(filename_or_obj, chunks=corrected_chunks, **kwargs)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/uxarray/core/api.py:531, in _get_grid(grid_filename_or_obj, chunks, chunk_grid, use_dual, grid_kwargs, **kwargs)
528 grid_kwargs["return_chunks"] = True
530 # Create a Grid
--> 531 return open_grid(grid_filename_or_obj, use_dual=use_dual, **grid_kwargs)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/uxarray/core/api.py:130, in open_grid(grid_filename_or_obj, chunks, use_dual, **kwargs)
126 grid = Grid.from_dataset(grid_filename_or_obj, use_dual=use_dual)
128 else:
129 # Attempt to use Xarray directly for remaining input types
--> 130 grid_ds = _open_dataset_with_fallback(
131 grid_filename_or_obj, chunks=grid_chunks, **kwargs
132 )
133 grid = Grid.from_dataset(grid_ds, use_dual=use_dual)
135 # Return the grid (and chunks, if requested) in a consistent manner.
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/uxarray/core/utils.py:38, in _open_dataset_with_fallback(filename_or_obj, chunks, **kwargs)
34 except Exception:
35 # If it fails, use the "netcdf4" engine as backup
36 # Extract engine from kwargs to prevent duplicate parameter error
37 engine = kwargs.pop("engine", "netcdf4")
---> 38 return xr.open_dataset(filename_or_obj, engine=engine, chunks=chunks, **kwargs)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/api.py:607, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, create_default_indexes, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
595 decoders = _resolve_decoders_kwargs(
596 decode_cf,
597 open_backend_dataset_parameters=backend.open_dataset_parameters,
(...) 603 decode_coords=decode_coords,
604 )
606 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 607 backend_ds = backend.open_dataset(
608 filename_or_obj,
609 drop_variables=drop_variables,
610 **decoders,
611 **kwargs,
612 )
613 ds = _dataset_from_backend_dataset(
614 backend_ds,
615 filename_or_obj,
(...) 626 **kwargs,
627 )
628 return ds
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:771, in NetCDF4BackendEntrypoint.open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, mode, format, clobber, diskless, persist, auto_complex, lock, autoclose)
749 def open_dataset(
750 self,
751 filename_or_obj: T_PathFileOrDataStore,
(...) 768 autoclose=False,
769 ) -> Dataset:
770 filename_or_obj = _normalize_path(filename_or_obj)
--> 771 store = NetCDF4DataStore.open(
772 filename_or_obj,
773 mode=mode,
774 format=format,
775 group=group,
776 clobber=clobber,
777 diskless=diskless,
778 persist=persist,
779 auto_complex=auto_complex,
780 lock=lock,
781 autoclose=autoclose,
782 )
784 store_entrypoint = StoreBackendEntrypoint()
785 with close_on_error(store):
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:529, in NetCDF4DataStore.open(cls, filename, mode, format, group, clobber, diskless, persist, auto_complex, lock, lock_maker, autoclose)
525 else:
526 manager = CachingFileManager(
527 netCDF4.Dataset, filename, mode=mode, kwargs=kwargs, lock=lock
528 )
--> 529 return cls(manager, group=group, mode=mode, lock=lock, autoclose=autoclose)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:429, in NetCDF4DataStore.__init__(self, manager, group, mode, lock, autoclose)
427 self._group = group
428 self._mode = mode
--> 429 self.format = self.ds.data_model
430 self._filename = self.ds.filepath()
431 self.is_remote = is_remote_uri(self._filename)
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:538, in NetCDF4DataStore.ds(self)
536 @property
537 def ds(self):
--> 538 return self._acquire()
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/netCDF4_.py:532, in NetCDF4DataStore._acquire(self, needs_lock)
531 def _acquire(self, needs_lock=True):
--> 532 with self._manager.acquire_context(needs_lock) as root:
533 ds = _nc4_require_group(root, self._group, self._mode)
534 return ds
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/contextlib.py:141, in _GeneratorContextManager.__enter__(self)
139 del self.args, self.kwds, self.func
140 try:
--> 141 return next(self.gen)
142 except StopIteration:
143 raise RuntimeError("generator didn't yield") from None
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:207, in CachingFileManager.acquire_context(self, needs_lock)
204 @contextmanager
205 def acquire_context(self, needs_lock: bool = True) -> Iterator[T_File]:
206 """Context manager for acquiring a file."""
--> 207 file, cached = self._acquire_with_cache_info(needs_lock)
208 try:
209 yield file
File ~/checkouts/readthedocs.org/user_builds/uxarray/conda/1439/lib/python3.14/site-packages/xarray/backends/file_manager.py:225, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
223 kwargs = kwargs.copy()
224 kwargs["mode"] = self._mode
--> 225 file = self._opener(*self._args, **kwargs)
226 if self._mode == "w":
227 # ensure file doesn't get overridden when opened again
228 self._mode = "a"
File src/netCDF4/_netCDF4.pyx:2521, in netCDF4._netCDF4.Dataset.__init__()
File src/netCDF4/_netCDF4.pyx:2158, in netCDF4._netCDF4._ensure_nc_success()
FileNotFoundError: [Errno 2] No such file or directory: '/glade/work/jpan/uxazim_demo/ne120np4_pentagons_100310.nc'
Step 2.1: Visualize the raw surface pressure field#
Because we are only interested in a single tropical cyclone, we can subset a region of the global field using a bounding circle to speed up plotting.
# Use a bounding circle to select faces whose centers lie
# within 5 great-circle degrees of the central point.
tcps = tcds["PS"].subset.bounding_circle((clon, clat), 5)
# Setting dynamic=True allows for quick, adaptive rendering as you zoom/pan.
# It also preserves the original mesh faces in the plot.
ps_plt = tcps.plot(
cmap="inferno", periodic_elements="split", title="Surface pressure", dynamic=True
)
ps_plt
Step 2.2: Compute the azimuthal mean of the 3D fields#
args = ((clon, clat), 3, 0.25)
azim_mean_T = tcds["T"].azimuthal_mean(*args)
azim_mean_Z = tcds["Z3"].azimuthal_mean(*args)
azim_mean_T
Step 2.3: Plot the TC radial profile#
The contour plot below shows the warm core and low pressure of the TC. After taking the azimuthal average, we subtract the value at the outermost radius to obtain an approximate anomaly relative to the ambient environment.
# Subtract the value at the outer radius
Tpert = azim_mean_T - azim_mean_T.isel(radius=-1)
Zpert = azim_mean_Z - azim_mean_Z.isel(radius=-1)
plt.contour(
azim_mean_Z["radius"],
tcds["plev"] / 100,
Zpert,
levels=np.arange(-500, 501, 50),
colors="black",
)
plt.contourf(
azim_mean_T["radius"],
tcds["plev"] / 100,
Tpert,
levels=np.arange(-10, 11, 2),
cmap="bwr",
extend="both",
)
plt.xlabel("distance from center [°]")
plt.ylim(1000, 100)
plt.yscale("log")
plt.ylabel("Pressure [hPa]")
plt.colorbar(label="T anomaly [K]")